Crucial in the development of joint tools, is the set-up of a reliable and curated DNA sequence library together with preservation of taxonomically validated vouchers. Linking DNA sequences and existing taxonomy (species names) is one of the strengths of GEANS, as it is important that data and results from traditional monitoring surveys can still be used after the implementation of genetic methods.
The focus of our reference library is on macrobenthic species of the North Sea, and more specifically those that occurred in our pilot study areas. It is a compilation of new species collected and sequenced during GEANS, quality checking and validating non-public and public COI sequences in databases kept at the partners institutions at the start of GEANS (e.g. Raupach et al. 2015; Barco et al. 2016; Laakman et al. 2016; Beermann et al. 2018; Rossel et al. 2020), and addition of reliable sequences from GenBank and BOLD.
This resulted in an open and well-curated DNA sequence reference library for macrobenthic species of the North Sea that contains 4005 COI sequences and that is hosted on BOLD. A total of 1947 sequences are new barcode records and have been additionally deposited in Genbank, while 2058 pre-existing sequences have been validated. A barcode for 89 species found in the North Sea was given for the first time. We are not covering all North Sea species, but it contains at least 712 species (including 69 NIS species), which is ca 32% of North Sea macrobenthos. Arthropoda is the most well represented taxon in number of sequences and species, followed by Mollusca (Fig. 1). Annelida, although recorded in a low number of sequences, are well represented with species in the list. Among all groups, Echinodermata sequences have the highest barcode coverage (56% when compared to the whole North Sea fauna).
The library is available at dx.doi.org/10.5883/DS-GEANS1
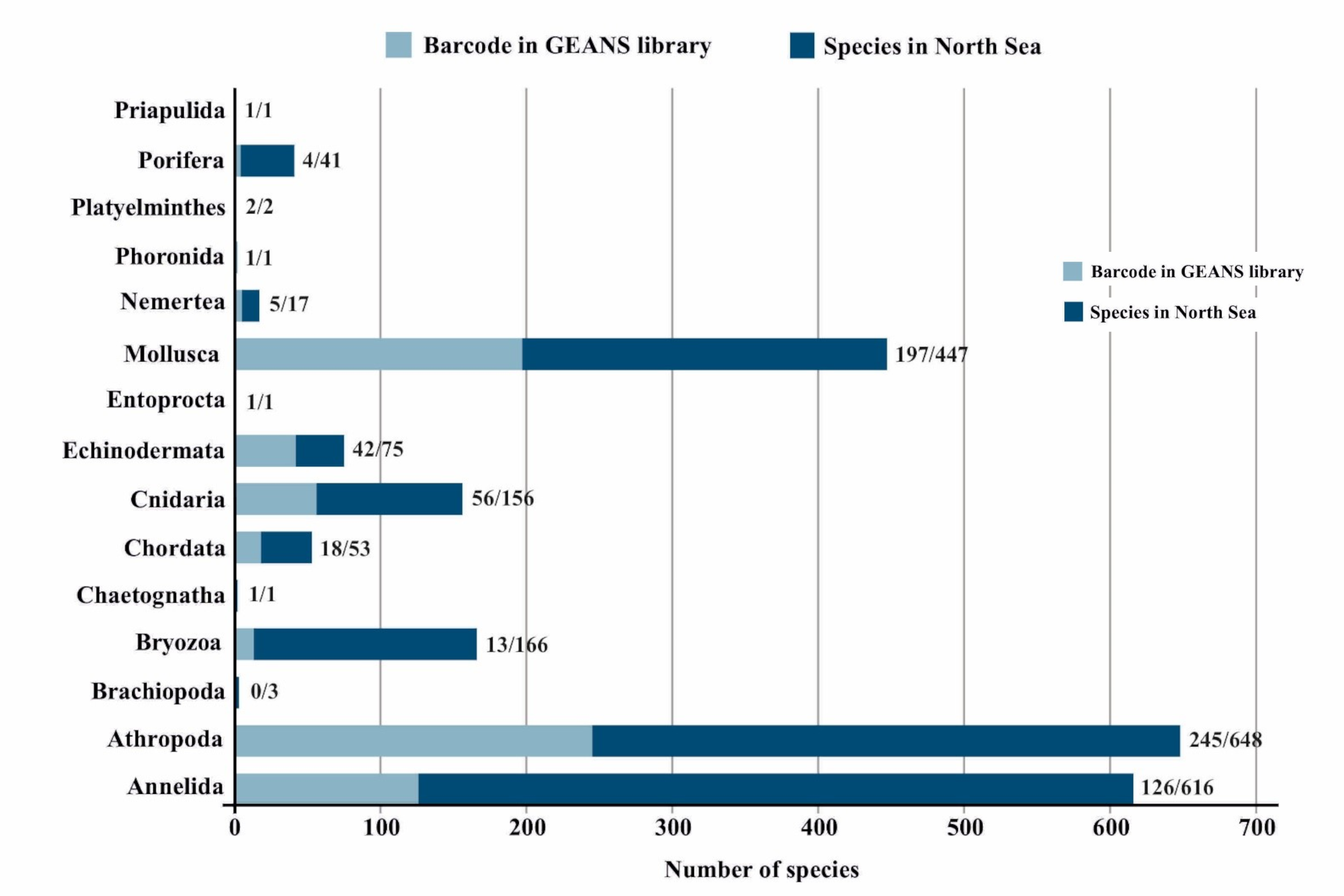
Figure 1: Barcode coverage of marine macrobenthic species of the North Sea in the GEANS DNA reference library. Numbers on bars refer to the species barcoded in comparison to the North Sea species.
References
Barco A, Raupach MJ, Laakmann S, Neumann H, Knebelsberger T. (2016) Identification of North Sea molluscs with DNA barcoding. Molecular Ecology Resources 16(1): 288-97. https://doi.org/10.1111/1755-0998.12440
Beermann J, Westbury MV, Hofreiter M, Hilgers L, Deister F, Neumann H, Raupach MJ (2018) Cryptic species in a well-known habitat: applying taxonomics to the amphipod genus Epimeria (Crustacea, Peracarida). Scientific Reports 8: 6893. https://doi.org/10.1038/s41598-018-25225-x
Laakmann S., Boos K., Knebelsberger T. Raupach MJ, Neumann H (2017) Species identification of echinoderms from the North Sea by combining morphology and molecular data. Helgoland Marine Research 70, 18. https://doi.org/10.1186/s10152-016-0468-5
Raupach MJ, Barco A, Steinke D, Beermann J, Laakmann S, Mohrbeck I, Neumann H, Kihara TC, Pointer K, Radulovici A, Segelken-Voigt A, Wesse C, Knebelsberger (2015) The Application of DNA Barcodes for the Identification of Marine Crustaceans from the North Sea and Adjacent Regions. PLoS ONE 10(9): e0139421. https://doi.org/10.1371/journal.pone.0139421
Rossel S, Raupach MJ (2020) First insights into the phylogeography and demographic history of the common hermit crab Pagurus bernhardus (Linnaeus, 1758) (Decapoda: Anomura: Paguridae) across the Eastern Atlantic and North Sea. Journal of Crustacean Biology, 40(4): 435–449. https://doi.org/10.1093/jcbiol/ruaa026